Using big data to target gene mutations in cancer tumor cells
Researchers are leveraging molecular profiling data to better understand certain cancers and uncover new treatments.

A single domino can send things in a new direction, much like how a gene mutation can initiate a chain reaction in cells and lead to cancer. University of Arizona Health Sciences researchers are using big data to target a particularly challenging gene mutation called KRAS.
There are many potential causes of cancer, from food and the environment to trauma and infection. When it comes to genetics, there is one gene mutation that researchers have linked to more than 20% of lung cancers, 40% of colorectal cancers and 90% of pancreatic cancers. The gene in question, KRAS, is one of the most common “oncogenes,” mutated genes that have the potential to cause cancer.
At the University of Arizona Health Sciences, researchers are applying vast amounts of data in an effort to learn more about this mutation, its variants and any correlating factors that might help them treat patients.
“We are leveraging a massive amount of data to stratify and profile patients, with and without KRAS mutations, in the hopes of finding areas where existing medications might be effective or reasons why some treatments meet resistance,” said Ritu Pandey, PhD, associate research professor in the Department of Cellular and Molecular Medicine in the College of Medicine — Tucson and associate director of Translational Clinical Bioinformatics in the UArizona Health Science s Center for Biomedical Informatics and Biostatistics. “Every patient having their own treatment pattern based on their own genetic makeup is the perfect example of precision medicine.”
What is a KRAS mutation?
The interaction between genes, proteins, and receptors is vital for maintaining life. Genes hold the recipe for making proteins, which do much of the work within a cell. These proteins talk to receptors to manage different tasks.
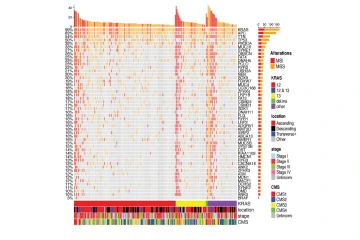
This figure shows characteristics of tumors in KRAS mutated colon cancer patients. Each column is a patient and rows are genes co-mutated along with KRAS.
In normal cells, the KRAS gene contains the instructions to make a protein called K-Ras, which acts like an on-off switch to regulate cell growth and proliferation. When the KRAS gene is mutated, the K-Ras protein becomes stuck in the “on” position, leading to the over multiplication of cells and, in some cases, cancer growth.
The K-Ras protein has long been considered an impossible target for medications because its slick surface gives drug molecules little to connect to take effect. In other words, the molecule lacks drug-binding sites. K-Ras protein mutations have several mutation subtypes, and there is no universal solution to target them, although there have been recent advancements in this area.
“There is a breakthrough where a drug has been found that can target a very specific kind of KRAS mutation called KRAS G12C, but nobody knows what to do with all the other variations as of yet,” Pandey said. “Learning more about those variants and finding solutions for them is the impetus behind our current research.”
Pandey and her collaborators are categorizing patient tumors into groups based on DNA, RNA and protein data to analyze and extract meaningful insights from the information. The highly refined and detailed research on KRAS is only possible thanks to data stored by the molecular profiling company Caris Life Sciences. The data is collected from the tumors of cancer patients across the United States, including at Banner – University Medical Center Tucson.
How KRAS mutations cause problems
In much the same way a KRAS mutation can drive cancer, so can a mutation in the epidermal growth factor receptor, or EGFR, gene. This causes an error in the EGFR protein function, which can lead to surplus cell growth.
In some cases, anti-EGFR treatments can slow or stop excess growth by inhibiting the EGFR protein, but this usually does not work for people who also have a KRAS-positive cancer.

Ritu Pandey, PhD, specializes in data driven research. She investigates molecular events using computational analysis and translates datasets into insights that advance the understanding of therapeutic targets.
“A faulty K-Ras protein keeps the EGFR signaling pathway active, even if the EGFR protein is inhibited,” Pandey said.
To visualize this, imagine a bustling factory where the EGFR and K-Ras proteins both work as supervisors of an assembly line. EGFR is one of the executives whose job is to receive signals from outside the factory and relay orders to increase or decrease production when needed. K-Ras is the foreman, working on the floor to direct some of the workers further down the line.
EGFR and K-Ras usually conduct their managerial duties in tandem. When a KRAS mutation occurs, however, the K-Ras foreman suddenly decides to tell its workers to ignore the EGFR executive’s prior instructions and ramp up production. This is how the on-off switch gets stuck in the “on” position and why drugs that inhibit EGFR don’t have an effect when there is a KRAS mutation.
KRAS’s interference with this otherwise effective therapy is also something Pandey’s research is scrutinizing. KRAS-mutated cells tend to exhibit differences in treatment outcome in different tumor types and in patients with the same tumor type, even with the same KRAS mutation subtype.
“We need to learn more about these processes to find out where in the assembly line we can come in with a treatment and make a correction or find an alternative mechanism to target the cell,” she said. “This requires going deep into genomics data and analyzing it to find any clues or signals that exist and any change in the molecular landscape in certain scenarios. It could be a combination of co-occurring mutations or other transcriptional changes in the mutated cells.”
Solutions for KRAS cancers and beyond
Pandey’s work would not be possible without Nirav Merchant, MS, director of the Data Science Institute, interim director of the Center for Biomedical Informatics and Biostatistics and leader of the Health Analytics Powerhouse, a UArizona Health Sciences strategic initiative.

Nirav Merchant, MS, leads the UArizona Health Sciences strategic initiative Health Analytics Powerhouse. This initiative supports many research projects by keeping research data secure while maximizing the benefits of advanced data collection and analysis techniques
“We’ve stood up several technologies that allow for secure data collection and storage at the university. This is a prerequisite for the kind of research Pandey is conducting,” Merchant said. “It is very exciting to see how our efforts have enabled such a powerful and potentially transformative project.”
To get the most out of the data, Pandey is working with UArizona Cancer Center oncologists, including Aaron Scott, MD, co-leader of the Clinical and Translational Oncology Program and associate professor in the College of Medicine—Tucson. He helps analyze the data to find clinically relevant information worthy of further examination.
“We know some people with KRAS-mutated colorectal cancer will have a poor response to chemotherapy, but right now, we cannot predict which one of those patients may respond better than others,” he said. “We will be able to refine and sort the different types of KRAS mutations along with other cellular changes that might contribute to better or worse responses to treatment. Ultimately, this could lead to the development of improved outcomes for patients with KRAS mutant colorectal cancer.”

Aaron Scott, MD, specializes in cancers of the gastrointestinal tract, which include colon, esophageal, stomach and pancreatic cancers. His particular interests are in clinical trial development and laboratory research investigating new therapies to improve outcomes.
Scott says this level of precision medicine will be a key to uncovering answers to questions that have long puzzled oncologists.
“Understanding how KRAS mutations differ, especially depending on primary tumor location in the colon, will also be an important to better understand the cancer biology and future treatment strategies,” he added.
For decades, KRAS mutations have puzzled researchers. Pandey’s unique application of big data could reveal paths toward new understandings, improved therapies and more precise treatment plans. The technological framework can also be shared across the university.
“She has built the scaffolding for improving our understanding of how KRAS mutant cancers behave, which in turn may lead to more effective treatments in the future,” Scott said of Pandey. “The KRAS mutation story is an example of how we are using precision medicine to improve our understanding of the biology of KRAS mutant cancers. The potential impact of this program’s development holds promise to be able to apply this line of research to other biological markers of cancer, too.”
Our Experts
Ritu Pandey, PhD
Associate Research Professor, Cellular and Molecular Medicine, College of Medicine – Tucson
Associate Director, Translational Clinical Bioinformatics, Center for Biomedical Informatics and Biostatistics, UArizona Health Sciences
Aaron J. Scott, MD
Associate Professor, Medicine, College of Medicine – Tucson
Co-Leader, Clinical and Translational Oncology Program, UArizona Cancer Center
Nirav Merchant, MS
Director, Cyber Innovation, Office of Research, Innovation and Impact
Director, Data Science Institute, Office of Research, Innovation and Impact
Interim Director, Center for Biomedical Informatics and Biostatistics, UArizona Health Sciences
Co-Principle Investigator, CyVerse
Member, BIO5 Institute
Contact
Brian Brennan
University of Arizona Health Sciences Office of Communications
520-621-3510, brianbrennan@arizona.edu

